Saving McCarty Woods
Written by: Roxanne Hoorn The Florida Museum of Natural History (FM) is promoting biodiversity and conservation efforts around the world, … Continue
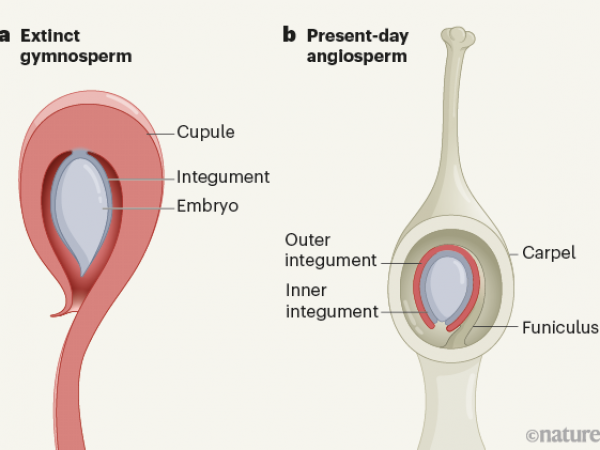
Ancient seeds spill secrets about the evolution of flowering plants.
Soltis, D. E. 2021. Ancient seeds spill secrets about the evolution of flowering plants. Nature, doi: 10.1038/d41586-021-01347-7. Nature Publishing Group. … Continue
Evolution of rapid blue‐light response linked to explosive diversification of ferns in angiosperm forests.
Cai, S., Y. Huang, F. Chen, X. Zhang, E. Sessa, C. Zhao, D. B. Marchant, D. Xue, G. Chen, F. … Continue
Spatial phylogenetics of butterflies in relation to environmental drivers and angiosperm diversity across North America.
Earl, C., M. W. Belitz, S. W. Laffan, V. Barve, N. Barve, D. E. Soltis, J. M. Allen, P. S. … Continue
Biogeography and habitat evolution of Saxifragaceae, with a revision of generic limits and a new tribal system.
Folk, R. A., R. L. Stubbs, N. J. Engle-Wrye, D. E. Soltis, and Y. Okuyama. 2021. Biogeography and habitat evolution … Continue
A new, simple, highly scalable, and efficient protocol for genomic DNA extraction from diverse plant taxa.
Mavrodiev, E. V., C. Dervinis, W. M. Whitten, M. A. Gitzendanner, M. Kirst, S. Kim, T. J. Kinser, P. S. … Continue
Examination of Reticulate Evolution Involving Haageocereus and Espostoa.
Arakaki, M., P. Speranza, P. S. Soltis, and D. E. Soltis. 2021. Examination of Reticulate Evolution Involving Haageocereus and Espostoa. … Continue
Green giant—a tiny chloroplast genome with mighty power to produce high-value proteins: history and phylogeny.
Daniell, H., S. Jin, X.-G. Zhu, M. A. Gitzendanner, D. E. Soltis, and P. S. Soltis. 2021. Green giant—a tiny … Continue
High-throughput methods for efficiently building massive phylogenies from natural history collections.
Folk, R. A., H. R. Kates, R. LaFrance, D. E. Soltis, P. S. Soltis, and R. P. Guralnick. 2021. High-throughput … Continue
Polyploidy: an evolutionary and ecological force in stressful times.
Van de Peer, Y., T.-L. Ashman, P. S. Soltis, and D. E. Soltis. 2021. Polyploidy: an evolutionary and ecological force … Continue